1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
| import cv2
import numpy as np
import matplotlib.pyplot as plt
import pydicom as pdicom
import os
import glob
import pandas as pd
import scipy.ndimage
from skimage import measure, morphology
from mpl_toolkits.mplot3d.art3d import Poly3DCollection
INPUT_FOLDER = './'
patients = os.listdir(INPUT_FOLDER)
patients.sort()
lstFilesDCM = []
def load_scan2(path):
for dirName, subdirList, fileList in os.walk(path):
for filename in fileList:
if ".dcm" in filename.lower():
lstFilesDCM.append(os.path.join(dirName, filename))
return lstFilesDCM
first_patient = load_scan2(INPUT_FOLDER)
print (lstFilesDCM[0])
RefDs = pdicom.read_file(lstFilesDCM[0])
ConstPixelDims = (int(RefDs.Rows), int(RefDs.Columns), len(lstFilesDCM))
ConstPixelSpacing = (float(RefDs.PixelSpacing[0]), float(RefDs.PixelSpacing[1]), float(RefDs.SliceThickness))
x = np.arange(0.0, (ConstPixelDims[0]+1)*ConstPixelSpacing[0], ConstPixelSpacing[0])
y = np.arange(0.0, (ConstPixelDims[1]+1)*ConstPixelSpacing[1], ConstPixelSpacing[1])
z = np.arange(0.0, (ConstPixelDims[2]+1)*ConstPixelSpacing[2], ConstPixelSpacing[2])
ArrayDicom = np.zeros(ConstPixelDims, dtype=RefDs.pixel_array.dtype)
for filenameDCM in lstFilesDCM:
ds = pdicom.read_file(filenameDCM)
ArrayDicom[:, :, lstFilesDCM.index(filenameDCM)] = ds.pixel_array
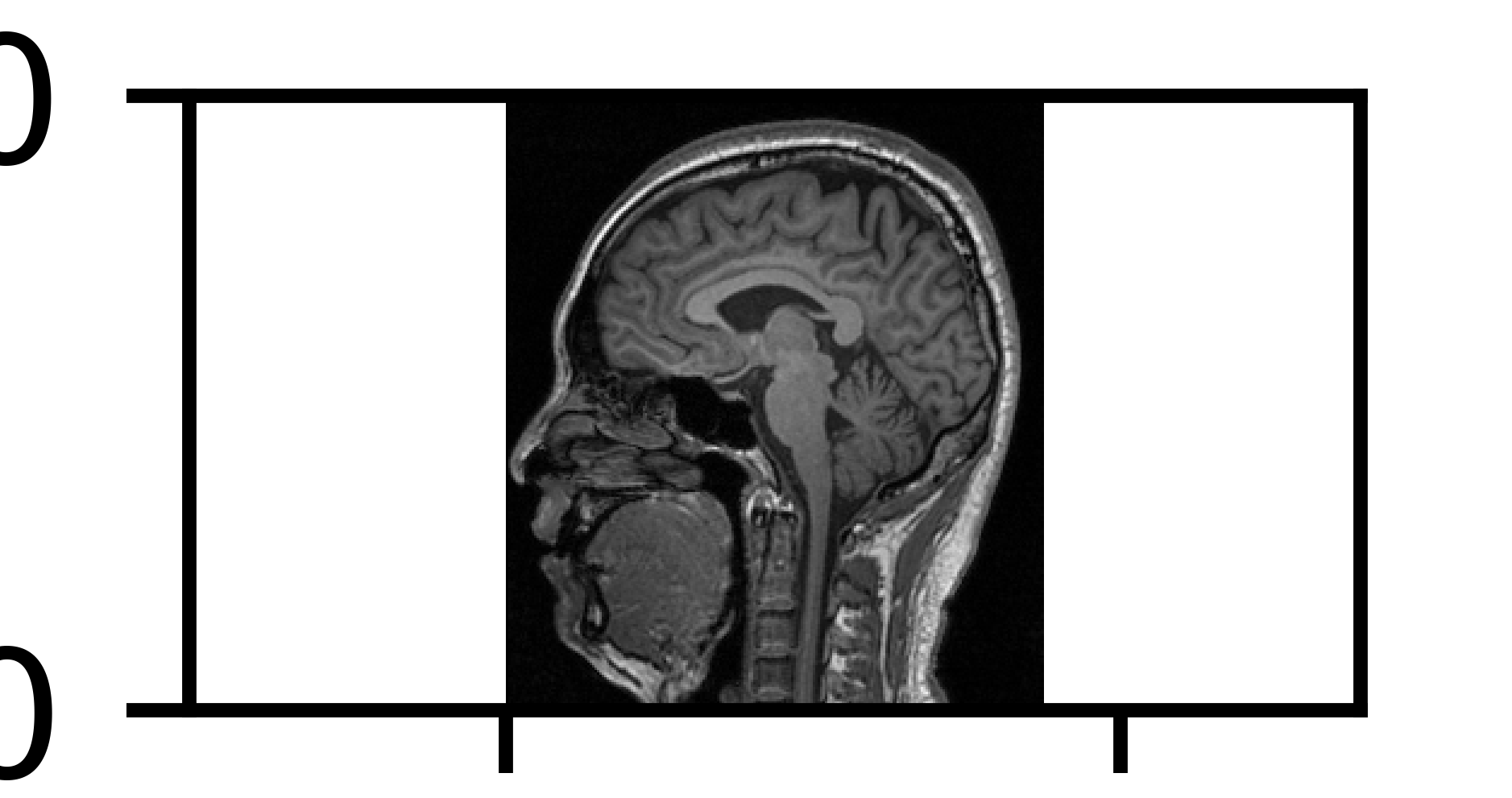
plt.figure(dpi=1600)
plt.axes().set_aspect('equal', 'datalim')
plt.set_cmap(plt.gray())
plt.pcolormesh(y, x, np.flipud(ArrayDicom[:, :, 80]))
plt.show()
|